What is KEGG? A Comprehensive Guide to the Kyoto Encyclopedia of Genes and Genomes
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a comprehensive bioinformatics resource that connects genomic information with biological functions through a collection of curated databases. These include KEGG PATHWAY for visualizing metabolic and signaling pathways, KEGG ORTHOLOGY (KO) for function-based gene annotation across species, KEGG MODULE for identifying functional units within pathways, KEGG LIGAND for biochemical compounds and reactions, and KEGG BRITE for hierarchical classification. KEGG plays a crucial role in functional genomics, disease research, drug discovery, and metagenomics. It enables researchers to annotate genes, predict functions, and map biological processes using tools like KEGG Mapper and KAAS, making it a vital platform in systems biology and bioinformatics analysis.
In modern biology and bioinformatics, one of the most important challenges is making sense of large-scale genomic data. With the rapid development of high-throughput sequencing technologies, researchers now generate vast amounts of genetic information. However, understanding the biological meaning of these data requires specialized tools. One of the most widely used resources for this purpose is KEGG — the Kyoto Encyclopedia of Genes and Genomes.
KEGG is a comprehensive database resource designed to link genomic information with higher-level systemic functions. It plays a central role in functional annotation, pathway analysis, drug discovery, and more. In this blog, we’ll explore what KEGG is, how it works, and why it matters in life sciences research.
What is KEGG?
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of manually curated databases and bioinformatics tools that integrate genomic, chemical, and functional information. Developed by Dr. Minoru Kanehisa and his team in Japan, KEGG helps scientists understand biological systems through molecular-level interactions.
It is widely used for:
- Annotating genes and proteins
- Analyzing metabolic and signaling pathways
- Exploring disease mechanisms
- Predicting drug-target interactions
By organizing biological knowledge into pathway maps and functional hierarchies, KEGG provides a visual and computational framework for interpreting complex biological data.
Core Components of KEGG
KEGG is composed of multiple interconnected databases. Each serves a specific role in biological data integration:
1. KEGG PATHWAY
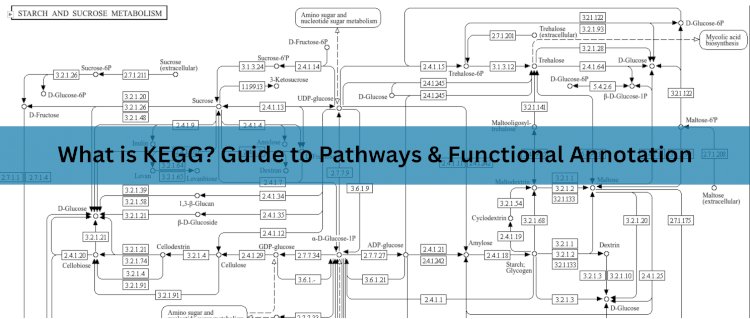
This is the most well-known part of KEGG. It includes pathway maps that represent:
- Metabolic pathways
- Genetic and environmental information processing
- Cellular and organismal processes
- Human diseases and drug development
These maps help visualize how genes and proteins work together in complex biological systems. For example, the glycolysis pathway shows the sequence of enzyme-catalyzed reactions involved in breaking down glucose to produce energy.
2. KEGG GENES and KEGG ORTHOLOGY (KO)
KEGG contains genome information from thousands of organisms. Genes are grouped into functional sets called orthologs, which are assigned KO (KEGG Orthology) numbers. KO identifiers allow researchers to connect genes across species based on their functional roles.
3. KEGG MODULE
Modules are smaller functional units that represent key steps in pathways or biological processes, such as a protein complex or a metabolic sub-pathway. They are useful for reconstructing pathways and identifying missing genes in incomplete genomes.
4. KEGG BRITE
BRITE is a hierarchical classification system that organizes enzymes, proteins, compounds, and drugs into function-based categories. It allows users to explore relationships beyond individual pathways.
5. KEGG LIGAND
This component includes several sub-databases:
- KEGG COMPOUND – chemical compounds in biological systems
- KEGG REACTION – biochemical reactions
- KEGG ENZYME – enzyme classifications (EC numbers)
- KEGG DRUG – pharmaceutical drugs and targets
- KEGG GLYCAN – information on glycans
These databases are essential for understanding biochemical networks, metabolism, and drug action.
Scientific Importance of KEGG
KEGG plays a vital role in functional genomics and systems biology. By integrating gene and protein data with known pathways and reactions, KEGG helps researchers:
- Predict gene function in newly sequenced organisms
- Understand the molecular basis of diseases
- Design metabolic pathways for synthetic biology
- Identify potential drug targets
- Explore microbiome functions in metagenomics studies
For example, after RNA-seq analysis, researchers often use KEGG pathway enrichment analysis to determine which biological pathways are activated or suppressed under certain conditions.
KEGG in Bioinformatics Workflows
KEGG is often used in combination with other tools and techniques:
- KEGG Mapper: Visualizes gene expression or metabolic activity on pathway maps.
- KEGG Automatic Annotation Server (KAAS): Assigns KO numbers to genes in a new genome.
- KEGG REST API: Allows programmatic access to KEGG databases for custom analyses.
KEGG is also integrated into popular bioinformatics software such as:
- DAVID
- GSEA (Gene Set Enrichment Analysis)
- MEGAN (for metagenomics)
- HUMAnN (for microbial community profiling)
Real-World Applications of KEGG
1. Disease Research
KEGG PATHWAY includes disease-specific pathways such as cancer, Alzheimer’s, and diabetes. By mapping patient gene expression data to these pathways, researchers can uncover disease mechanisms and identify biomarkers.
2. Drug Discovery
KEGG DRUG and ENZYME databases allow scientists to find potential drug targets and understand how drugs interact with genes and proteins.
3. Microbial Genomics
In environmental microbiology and industrial biotechnology, KEGG MODULE is used to reconstruct metabolic capabilities of microbes from genomic or metagenomic data.
4. Agricultural Genomics
KEGG supports functional annotation in crops and livestock, aiding in trait discovery, breeding, and improving stress resistance.
Advantages of KEGG
- Manually curated and reliable
- Covers a broad range of species
- Visual pathway maps aid interpretation
- Integrated view of genes, proteins, and compounds
- Useful for both basic research and applied sciences
Hands-On Training in KEGG Annotation at IBRI Noida
The Indian Biological Sciences and Research Institute (IBRI), Noida is emerging as a leading center for life sciences education and skill development in India. As part of its commitment to fostering industry-ready professionals, IBRI offers hands-on training in KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation and pathway analysis. Through practical workshops and real-time bioinformatics projects, students and researchers gain direct experience in using KEGG tools for functional genomics, metabolic pathway reconstruction, and drug-target identification. This training equips participants with critical skills required in modern research labs and biotech companies, bridging the gap between academic knowledge and practical application in systems biology and bioinformatics.
Conclusion
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is an essential resource for anyone working in genomics, molecular biology, or systems biology. It bridges the gap between raw sequence data and biological understanding by organizing functional information into an accessible, visual format.
Whether you’re analyzing differential gene expression, annotating a genome, or developing a new drug, KEGG provides the tools and insights needed to connect data to biological meaning.