Sequencing Stage in the NGS Pipeline: Turning Libraries into Data

Sequencing Stage in the NGS Pipeline: Turning Libraries into Data
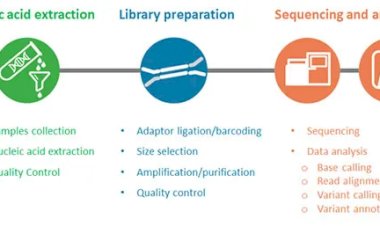
Welcome back to our series focused on the Next-Generation Sequencing (NGS) pipeline! In the previous blog, we explored the Library Preparation stage, where DNA or RNA fragments are processed into ready-to-sequence libraries. Now, it's time to delve into the Sequencing Stage—the pivotal step that transforms these libraries into raw data for analysis.
What is Sequencing in NGS?
Sequencing is the process of determining the precise order of nucleotides (A, T, G, and C) in the DNA or RNA fragments of your prepared library. This step leverages advanced technology to generate vast quantities of data, allowing researchers to uncover the genetic blueprint of an organism, identify mutations, or study gene expression patterns.
Why is Sequencing Important?
The sequencing step is crucial because it bridges the gap between prepared libraries and actionable data. Without this step, the insights locked in your samples would remain inaccessible. Modern sequencing platforms can generate billions of reads in a single run, enabling comprehensive studies of entire genomes, exomes, or transcriptomes.
The Sequencing Process: Step by Step
Loading the Library onto the Sequencer
Once the library is prepared, it is loaded onto a sequencing platform. Each fragment in the library is hybridized onto a solid surface or bead coated with complementary oligonucleotides. This ensures that the library fragments are immobilized and ready for sequencing.
Amplification (Optional, Platform-Specific)
Some platforms, such as Illumina’s, use bridge amplification to create clusters of identical DNA fragments. These clusters enhance the signal strength, ensuring accurate detection during sequencing. Other platforms, like PacBio or Oxford Nanopore, sequence single molecules directly, eliminating the need for amplification.
Sequencing-by-Synthesis or Alternative Techniques
Modern sequencing technologies use different principles to read the sequence of nucleotides. The two most common methods are:
Sequencing-by-Synthesis (SBS): Platforms like Illumina and MGI use SBS, where fluorescently labeled nucleotides are incorporated into the DNA strand, and a camera captures the emitted light to identify each nucleotide in real-time.
Nanopore-Based Sequencing: Oxford Nanopore devices detect changes in electrical conductivity as DNA or RNA passes through a nanopore.
Single-Molecule Real-Time (SMRT) Sequencing: Used by PacBio, this method tracks DNA polymerase activity in real-time to determine the sequence.
Data Generation
The sequencer converts the detected signals (light, current changes, etc.) into digital data. This data is then stored as raw reads, typically in FASTQ format, which contains sequence information and corresponding quality scores.
Key Sequencing Technologies
1. Illumina Sequencing
Illumina sequencing is based on a method called sequencing by synthesis (SBS). DNA fragments are immobilized on a flow cell, where they are amplified into clusters. During the sequencing process, fluorescently labeled nucleotides are added one at a time. Each base incorporated into the DNA strand emits a specific fluorescence signal, which is captured by a camera. The signal is then used to determine the base sequence. Illumina technology is known for its high accuracy and throughput, making it one of the most widely used platforms.
2. Ion Torrent Sequencing
Ion Torrent sequencing works by detecting the release of hydrogen ions during DNA synthesis. DNA fragments are attached to beads, amplified, and deposited into wells on a microchip. As nucleotides are added to the DNA strands, the resulting release of hydrogen ions changes the pH in each well. These pH changes are measured and converted into digital base calls. This method is faster and more cost-effective for certain applications but can have lower accuracy in homopolymer regions.
3. Pacific Biosciences (PacBio) Sequencing
PacBio’s single-molecule real-time (SMRT) sequencing observes DNA polymerase activity as it synthesizes DNA in real time. DNA fragments are attached to wells called zero-mode waveguides (ZMWs), and fluorescently labeled nucleotides emit signals as they are incorporated. This technology excels in producing long reads, which are critical for resolving complex genomic regions and structural variations.
4. Oxford Nanopore Sequencing
Oxford Nanopore technology uses a unique approach where DNA or RNA molecules are passed through protein nanopores embedded in a membrane. Changes in electrical current as the molecules pass through the nanopore allow the sequence of bases to be determined. Nanopore sequencing is notable for its ability to produce ultra-long reads, offering unparalleled flexibility for genome assembly and real-time analysis.
Challenges in the Sequencing Step
Despite advancements, the sequencing step comes with its own set of challenges.
Accuracy is a significant concern, particularly in repetitive or GC-rich regions, which can lead to misreads. Different platforms vary in their error profiles, such as homopolymer errors in Ion Torrent or indel errors in long-read technologies.
Throughput is another critical factor; while platforms like Illumina excel in producing large volumes of short reads, balancing high throughput with long-read accuracy remains a challenge.
Additionally, data complexity from high-throughput sequencing requires robust bioinformatics tools for accurate interpretation. The quality of the initial library preparation also plays a critical role, as degraded or poorly prepared samples can reduce sequencing efficiency and data quality.
Advancements and Optimizations
The field of NGS sequencing is continually evolving with innovations that improve its efficiency, accuracy, and accessibility. Enhanced chemistries and reagents now allow for more precise base calling and reduced error rates.
Many platforms are integrating automation, reducing the need for manual intervention and minimizing the chances of errors. Real-time sequencing technologies, such as PacBio and Oxford Nanopore, have shortened turnaround times while enabling the study of epigenetic modifications alongside nucleotide sequences.
Furthermore, advances in multiplexing strategies allow simultaneous sequencing of multiple samples, increasing throughput and cost-efficiency.
Artificial intelligence (AI) is also playing a growing role in sequencing, assisting in error correction, data analysis, and sequence assembly.
Preparing for the Next Step: Data Analysis
Once sequencing is complete, the raw reads must undergo data preprocessing and analysis to extract meaningful insights. This includes tasks like quality control, alignment to a reference genome, and variant calling. These topics will be covered in upcoming blogs in this series.
Conclusion
The sequencing stage is the heart of the NGS pipeline, converting carefully prepared libraries into valuable raw data. With advancements in sequencing technologies, researchers can now explore genomes with unparalleled accuracy and depth. However, understanding the nuances of each platform and sequencing method is crucial to maximizing the potential of your experiments.
Stay tuned for the next blog in this series, where we’ll dive into Data Preprocessing and Quality Control—the essential steps following sequencing that ensure the reliability of your data!
Also Read: Next-Generation Sequencing (NGS): A Beginner’s Guide & Its Scope
https://www.ibri.org.in/blog/next-generation-sequencing-ngs-a-beginners-guide-its-scope
#nextgenerationsequencing #genomics #DNAsequencing #illuminasequencing #ibricourses
#bioinformatics #genomeanalysis #molecularbiology #biotechnology #librarypreparation

 Manju
Manju